Several Matlab functions have been written for help with non-ideal multicomponent simulations. The following table lists the functions that have been written and tested to date:
|
|
|
|
|
|
|
>> [t,ys]=bubptg(p,xs) |
|
|
|
>> buberg(3,amipa) |
|
|
|
>> [p,ys]=bubtpg(t,xs) |
|
|
|
>> buberg2(3,amipa2) |
|
|
|
>> gams=marg1(xs) |
|
|
|
>> minvle(Amarg) |
|
|
|
>> XG=terf1(amipa) |
The ternary sytem: acetone, methanol, isopropyl alcohol will be the example that we use to demonstrate the programs. Data for this system was reported by Freshwater and Pike in the J. Chemical Engineering Data 12 page 179 1967. Two tables of ternary data were presented for this system. In the first set, the pressure was fixed at 760 mmHg and in the second set the temperature was fixed at 55C. The data for each set stored in a text file that was then read into a Matlab session. The isobaric data was stored in a file called amipa. It looked like the following when listed in Matlab:
x1 x2 y1 y2 T in C p in mmHg
0.0360 0.5460 0.1050 0.6750 69.3000 760.0000
0.0930 0.5230 0.2390 0.5810 66.8400 760.0000
0.1360 0.2980 0.3350 0.3690 68.8300 760.0000
0.0370 0.1120 0.1210 0.1699 76.9700 760.0000
0.8708 0.0761 0.8866 0.0912 56.3200 760.0000
0.5459 0.2693 0.6680 0.2569 58.4500 760.0000
0.3555 0.1917 0.5780 0.2096 63.6500 760.0000
0.0520 0.0287 0.1606 0.0485 77.4100 760.0000
0.1324 0.2635 0.3218 0.3391 69.2300 760.0000
0.0950 0.4453 0.2306 0.5135 68.1800 760.0000
0.0601 0.8993 0.1300 0.8556 63.1000 760.0000
0.1231 0.6965 0.2469 0.6776 63.2700 760.0000
0.3948 0.4841 0.5346 0.4237 58.4300 760.0000
0.2635 0.3920 0.4576 0.3861 62.9500 760.0000
0.3089 0.1895 0.5465 0.2040 64.6500 760.0000
0.0820 0.5602 0.1955 0.6259 66.7900 760.0000
0.3167 0.4201 0.4923 0.4023 61.2900 760.0000
0.3732 0.5910 0.4982 0.4923 57.5400 760.0000
0.4062 0.5355 0.5315 0.4490 57.5600 760.0000
0.4089 0.0351 0.6596 0.0552 64.7300 760.0000
0.4854 0.1379 0.6768 0.1480 61.7700 760.0000
The program terf1 was used to convert this data into an array containg the liquid mols fractions of all the compounds followed by the liquid activity coefficients by the following definition:
where: yi is the vapor mol fraction of compound i
xi is the liquid mol fraction of compound i P is the total pressure and pi*(T) is the vapor pressure of compound i.
Here is a listing of the conversion program:

This program was used to create the array XG1 from the isobaric data set in amipa and the array XG2 from the isothemal data set in amipa2. Property data for the three compounds (including "Mass & Energy" balance data) must be set with start301 before it can be executed. Here is a listing from Matlab of XG1:
x1 x2 x3 gam1 gam2 gam3
0.0360 0.5460 0.4180 1.8890 1.0322 0.8961
0.0930 0.5230 0.3840 1.7991 1.0194 0.8880
0.1360 0.2980 0.5660 1.6191 1.0526 0.9086
0.0370 0.1120 0.8510 1.6747 0.9526 1.0295
0.8708 0.0761 0.0531 1.0089 1.6763 1.2796
0.5459 0.2693 0.1848 1.1280 1.2222 1.1251
0.3555 0.1917 0.4528 1.2615 1.1366 1.0236
0.0520 0.0287 0.9193 1.5609 1.0444 1.0441
0.1324 0.2635 0.6041 1.5776 1.0774 0.9586
0.0950 0.4453 0.4597 1.6286 1.0049 0.9947
0.0601 0.8993 0.0406 1.7087 1.0108 0.7933
0.1231 0.6965 0.1804 1.5756 1.0266 0.9290
0.3948 0.4841 0.1211 1.2490 1.1223 0.9542
0.2635 0.3920 0.3445 1.3786 1.0526 1.0217
0.3089 0.1895 0.5016 1.3288 1.0758 1.0380
0.0820 0.5602 0.3578 1.6717 1.0272 0.9477
0.3167 0.4201 0.2632 1.3032 1.0935 0.9724
0.3732 0.5910 0.0358 1.2690 1.1078 0.7667
0.4062 0.5355 0.0583 1.2430 1.1142 0.9654
0.4089 0.0351 0.5560 1.2085 1.5667 1.0666
0.4854 0.1379 0.3767 1.1506 1.2022 1.1049
Here is the function used to find the "best" parameters to fit the data in an array like XG1.

The following session shows a test of this program and the beginning of the execution of fmins to find the "best" values for the parameters to fit the isobaric data:
»global Amar gamf XG
»gamf='marg1';
»XG=XG1;
»minvle([0 0 0])
ans =
13.7054 0 0 0 <-- sum of abs errors is 13.7
ans = for all zeros in Aber
13.7054
»fmins('minvle',[0 0 0])
ans =
13.7054 0 0 0
ans =
13.7028 0.0003 0 0
ans =
13.7041 0 0.0003 0
....many iterations later.....
ans =
2.8785 0.6740 0.4880 -0.0639
ans =
2.8785 0.6740 0.4879 -0.0640
ans =
2.8785 0.6740 0.4880 -0.0639 <-- sum of abs errors is 2.88
ans = "best" values
0.6740 0.4880 -0.0639
»Amar1=ans
Amar1 =
0.6740 0.4880 -0.0639 <-- We will save these best values for
the isobaric data.
»XG=XG2;
»fmins('minvle',Amar1) <-- Using Amar1 as an initial guess for the
ans = isothermal set
4.4551 0.6740 0.4880 -0.0639
....many iterations later.....
ans =
3.5376 0.6588 0.6060 0.0370
ans =
0.6588 0.6060 0.0370 <-- Best for isothermal data
»Amar2=ans;
»XG=[XG1;XG2]; <-- Putting in both sets of data
»Amar3=fmins('minvle',(Amar1+Amar)/2)
ans =
7.0304 0.6664 0.5470 -0.0135
....many iterations later.....
ans =
7.0100 0.6582 0.5409 -0.0142
Amar3 =
0.6582 0.5409 -0.0142 <-- Best for combined data.
In order to see how well the Margules' activity coefficient model predicts the original data, we need a function to determine the bubble point of our fluid. This requires only a slight modification of the ideal bubble point function: bubpt.
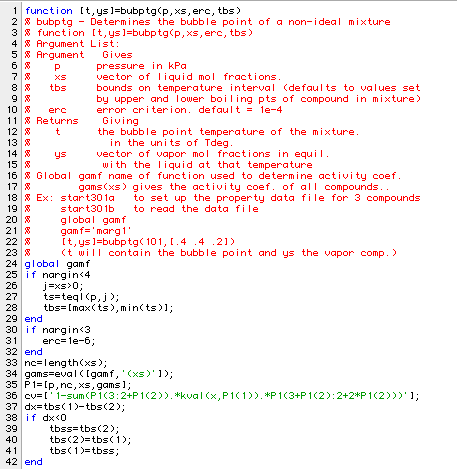

We will see how well we fit the first line of data listed in amipa:
»amipa(1,:)
ans =
0.0360 0.5460 0.1050 0.6750 69.3000 760.0000
»[t,ys]=bubptg(101.325,[.036 .546 .418])
t =
69.4051 <-- compared to 69.3
ys =
0.0984 0.6565 0.2451 <-- exp. values: 0.105 0.675 0.220
Our next program in this section of the notes, mechanizes the process of comparing the bubble points from the experiental data with those predicted by either the ideal program or one using an activity coefficient model such as Margules. Here is the program:
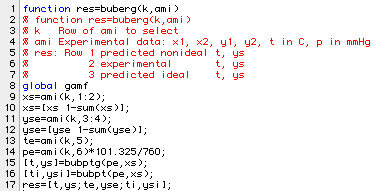
Executing buberg for the first line of data in amipa produces:
»buberg(1,amipa) ans = 69.4051 0.0984 0.6565 0.2451 <-- Margules predictions 69.3000 0.1050 0.6750 0.2200 <-- Experimental data 70.4860 0.0577 0.6840 0.2583 <-- Ideal prediction
Two additional programs were written to aid in comparing the isothermal data with the Magules model. The first program: bubtpg determines the pressure and vapor composition for known temperature and liquid composition. Here is a listing:
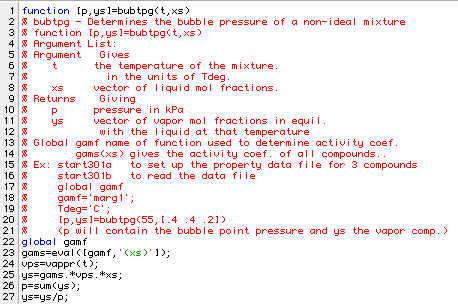
Here is a sample run using data in row 3 of amipa2:
»amipa2(3,:)
ans =
0.5372 0.3447 0.6480 0.3048 55.0000 695.9000
»[p,ys]=bubtpg(55,[0.5372 0.3447 0.1181])
p =
93.0284
ys =
0.6459 0.3091 0.0449 <-- compared to 0.648 0.3048 .0472
»p*760/101.325
ans =
697.7702 <-- compared to 695.9
The function buberg2 compares each isothermal data set just as buberg did for the isobaric data.
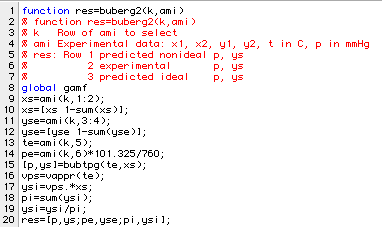
Here is what the program gives for the third row in amipa2:
»buberg2(3,amipa2) ans = 93.0284 0.6459 0.3091 0.0449 <-- Margules' prediction 92.7790 0.6480 0.3048 0.0472 <-- Experimental data 79.7977 0.6578 0.2962 0.0460 <-- Ideal prediction