HaeIII
from
Haemophilus aegytius
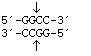
Blunt end
HindIII
from
Haemophilus influenzae Rd
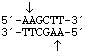
5' overhang sticky end
PvuI
from
Proteus vulgaris
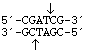
3' overhang sticky end
 Home
Home
The isolation, characterization, and commercial production of restriction enzymes represents a cornerstone of molecular biology. Three classes of these endonucleases are used:
The restriction enzymes cleave the phosphodiester bonds in each strand of double-stranded DNA:
Examples of class II restriction enzymes
|
HaeIII from |
|
Blunt end |
|
HindIII from |
|
5' overhang sticky end |
|
PvuI from |
|
3' overhang sticky end |
Restriction enzymes are obtained from many prokaryotes and about 1500 enzymes with known sequence recognition sites have been isolated. Naming these endonucleases follows a system proposed by Nathans and Smith. Each name contains at least one capital letter and two small letters followed by a Roman numeral. The letters are initials of the genus and species of origin and the number represents the number of enzymes discovered in the organism. (Historically the numeral identified the protein peak in which the enzyme eluted during chromatography.) Additional information may be added as a letter. For EcoRI, the R indicates the particular strain of E. coli.
Restriction enzymes from different organisms may recognize the same DNA sequence. If the enzymes recognize the same site and cleave at the same position, they are labeled isoschizomers. Ones that recognize the same site but cleave in different positions are heteroschizomers or neoschizomers.
|
Isoschizomer example |
||||||
|
SphI from |
|
BbuI from |
|
|||
|
Heteroisoschizomer or neoschizomer example |
||||||
|
SmaI from |
|
XmaI from |
|
|||
Iso- or heteroisoschizomers add flexibility to experimental design. Cost, methylation sensitivities, and types of "ends" are considerations as well as buffer conditions for optimal activity.
A few buffer conditions suit nearly all the restriction enzymes but no single buffer allows activity of every enzyme. Suppliers of enzymes always provide a reaction buffer (10x concentrate) that is optimum for the enzyme. Components of the 1x buffer usually are 10-100 mM Tris at pH 7.3 to 8.5, various levels of salts like KCl and NaCl (10 to 150 mM), 10 mM Mg(2+), 2 mM beta-mercaptoethanol. Sometimes 0.01% Triton- X100 (a detergent) and bovine serum albumin are included as a stabilizers. (Alternatively, swine skin gelatin can be used and offers the advantages that it is stable to autoclaving and costs about 1/15 as much as BSA.)
Since restriction enzymes can require different buffer conditions, some strategy must be used to do double digests. The preferred method is to simultaneously digest with both enzymes in a compatible buffer. This method can be used even if one enzyme is not fully active (e.g., 75% active). More of one enzyme can be added (e.g., 1 U of enzyme A + 1.33 U enzyme B) for equal cutting efficiency. There are limits to the excess enzyme due to increased glycerol in the reaction that can reduce specificity of some enzymes.
An alternative method is to digest with the "low salt" enzyme then add more buffer and the "high salt" enzyme to complete the digest. This obviously doubles the time required for digestion. In extreme cases the DNA can be precipitated after one digest and dissolved in the second digest buffer. Digests are carried out at 37 degrees C unless otherwise noted for the enzyme.
Non-specific or relaxed specificity cleavage or "star" activity can occur if non-optimal conditions are used. Conditions that encourage star activity include:
The state of the DNA also influences the ability of restriction enzymes to cut efficiently. "Dirty" DNA contaminated with protein, salts, and RNA is not cut by some enzymes. DNA from sources that methylate adenine or cytosine residues may not be digested (methylated sensitive, Table 6, pp. 126-134 Lab Fax or a catalogue appendix). Some strains of E. colimethylate the N(6) position of adenine in the sequence, GATC, and are denoted as dam . Others methylate the C(5) position of the internal cytosine in sequences of CCAGG, CCTGG and are designated as dcm. DNA from other sources (e.g., mammals, plants) can be methylated at other sites. Some restriction enzymes will not cut near these methylations if their recognition site overlaps with the methylation site.
Miscellaneous information on restriction enzymes
Copyright, Acknowledgements,
and Intended Use
Created by B. Beason (bbeason@rice.edu), Rice University, 6 February 2008
Updated 10 February 2008