 Home
Home
In both gel media, DNA is driven through the matrix by electric current. Smaller or more compact molecules pass through the matrix easier and migrate farther than large molecules. All linear DNA has the same charge per unit length and linear pieces migrate according to size. Plasmid DNA preparations contain three types of DNA conformations: linear, relaxed circular (or nicked) and supercoiled. Only the linear form can be used to estimate the size of the molecule. Usually, but not always, the supercoiled runs fastest, linear next, then the relaxed circular. A carefully prepared sample will be mostly supercoiled. The range of sizes separated in a gel is controlled by the % of agarose or %T of acrylamide in the gel.
Resolution Versus Matrix Concentration
|
Agarose |
Useful for Range of Linear dsDNA Molecules (kb) |
Acrylamide |
Useful for Range of Linear dsDNA Molecules (bp)* |
|
|
0.3 0.6 0.7 0.9 1.2 1.5 2.0 |
5 - 60 1 - 20 0.8 - 10 0.5 - 7 0.4 - 6 0.2 - 3 0.1 - 2 |
3.5 5.0 8.0 12.0 15.0 20.0 |
100 - 1000 75 - 500 50 - 400 35 - 250 20 - 150 5 - 100 |
The mobility is proportional to the voltage applied at low voltage but increasing voltage decreases the resolution of larger fragments of DNA. A general guideline for agarose gels in 1xTBE is 5V/cm maximum for resolving fragment lengths greater than 2 kb. The distance between the electrodes serves as the length in the calculation. Higher voltages increase the temperature of the gel causing increased band width and distortion of the lanes. The agarose can also melt, especially the low melting point agarose sometimes used when DNA is to be recovered from the gel. Voltages for acrylamide gels are generally twice the recommended voltage of the agarose gels but it is important to check manufacturer recommendations for the gel or for the electrophoretic equipment.
The mobility is also influenced by the choice of buffer systems. Besides the Tris Borate EDTA, pH 8.3 (TBE) buffer used in our experiments, a Tris Acetate EDTA buffer (TAE) is preferred by some. The TAE buffer shifts the range of resolution toward higher fragment lengths.
Denaturing gels can also be run to separate fragments as single stranded DNA. Gels can be run at high pH (30 mM NaOH) or in neutral buffers when glyoxal is added to the gel and the running buffer. Only the glyoxal system is suitable for RNA separations. Why is there a restriction of systems for RNA separations?
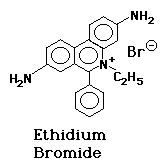
The nucleic acids are visualized with ethidium bromide (EtBr). This fluorescent dye, which contains a tricyclic planar group, intercalates between stacked base pairs of nucleotides and, in this environment, fluoresces when excited with ultraviolet light; the fixed position of the planar group and its close proximity to the bases causes dye bound to DNA to display increased fluorescent yield compared to free dye.
The size of linear fragments of DNA is determined by comparison
to standards in the same manner as protein molecular mass
is determined from SDS-PAGE. The log (# base pairs) is plotted
versus distance migrated or Rf value {Helling R.B., Goodman
H.M., and Boyer H.W. 1974. Analysis of endonuclease R-EcoRI
fragments of DNA from lambdoid bacteriophages and other viruses
by agarose-gel electrophoresis. J. Virol.14: 1235-1244}.
The molecular weight DNA markers used in our study are Quick-Load
1 kb DNA Ladder (New England Biolabs,
Catalog #N0468S). The ladder produces eleven DNA fragments
ranging in size from 500 to 10,000 base pairs. The ladder can
be used to quantitate the amount of DNA in a sample; the mass
of DNA in each band in the ladder has been calibrated.
The tracking dye combined with the DNA samples contains bromophenol blue and xylene cyanol for use in visually monitoring electrophoresis and glycerol to make the sample dense enough sink to the bottom of the well. The stock solution is designed to be diluted about six fold in the sample. Bromophenol blue runs about the same size as a linear double-stranded DNA molecule of 300 base pairs in length in 1X TBE on a gel of 1% agarose. In low percentage gels of 0.4% agarose, the dye can emulate a 1000bp fragment. Remember not to run this dye off the bottom of the gel when you are trying to analyze small fragments. Xylene cyanol runs about the same as a linear double-stranded DNA molecule of 4kb in a 1% agarose gel.
Copyright, Acknowledgements,
and Intended Use
Created by B. Beason (bbeason@rice.edu), Rice University, 21 November 2007
Updated 31 January 2008