 Home
Home
If we begin with certainties, we shall end in doubts; but if we begin with doubts,
and are patient in them, we shall end in certainties.
Francis Bacon
Day 1
***Prepare for the course by familiarizing yourself with the Background on
Microarrays.***
Foreword
- The detailed notes about each procedure are presented just before the protocol. These notes are meant to be read before each lab period. Please make special note of any points indicated with a stop sign or a hand. Some of the protocols are direct instructions from the manufacturer of the kit. Some notes are presented in a second section that simplify or customize the application for the teaching lab situation. You will have to read through all the material and compile detailed instructions in your notebook that incorporate all the special instructions. This approach is meant to give you experience in deciphering and assembling procedures.
- Recipes for any reagents required in a protocol are provided for your reference or for the instructor. All stock solutions will be provided.
Isolation of Total RNA from Arabidopsis thaliana
 High quality RNA is critical to the success of microarray experiments. Degraded or contaminated RNA cannot be efficiently reverse transcribed or labeled
and, therefore, will not produce good hybridization signals.
High quality RNA is critical to the success of microarray experiments. Degraded or contaminated RNA cannot be efficiently reverse transcribed or labeled
and, therefore, will not produce good hybridization signals.
In preparation for today's lab, the following were
performed by Dr. Elizabeth Eich (formerly McCormack):
- Total RNA was isolated from control and experimental Arabidopsis using TRIzol
Reagent (pdf from Invitrogen). Trizol is a monophasic
mixture of phenol and guanidine isothiocyanate; during
sample homogenizaiton, RNA integrity is maintained without
solubilization or precipitation of other cellular components.
Here's a shortened version of the RNA prep that is more specific
to technique for plants:
- Grow wild type and mutant plants to desired stage in
plant culture media.
- Collect seedlings and freeze in liquid N2.
- Use pestles to grind tissue in liquid N2.
- Add 200 µl of TRIzol for approximately 100 µl of powdered
tissue.
- Incubate at room temp. for 5 minutes.
- Add 40 µl chloroform, mix, and incubate at room temp. 2-3
minutes.
- Centrifuge at 12,000 x g for 15 minutes at 4°C.
- Wash pellet with 200 µl 70% EtOH (RNase free, made with DEPC
water).
- Centrifuge at 7500 x g for 5 minutes at 4°C.
- Air dry pellet.
- Resuspend RNA in DEPC water.
- Store RNA at -80°C.
- The concentration of RNA was determined by measuring the
absorbance at 260 nm; A260/A280 was determined to give an estimate
of the purity of RNA with respect to protein contaminants.
- The integrity of the 26S and 18S ribosomal bands were verified
by agarose gel electrophoresis; the ribosomal bands should
appear as sharp bands:
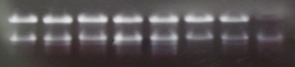
*Sample agarose gel
Genisphere 3DNA Array 900™ Expression Array Detection Kit
*Download the complete Array 900 Protocol in PDF format.
Do NOT print the entire manual--just read pages 2-15 and 20-21.
The 3DNA Array 900 kit is designed for arrays printed with PCR products (cDNA) or oligonucleotides.
First, total RNA is reverse transcribed using a special RT primer. Next, the cDNA and the fluorescent 3DNA reagent are hybridized to the array in succession; the 3DNA reagent contains a "capture sequence" that is complementary to a sequence on the 5' end of the RT primer.
Step 1: cDNA Synthesis from Total RNA (Reverse Transcription Reaction)
 Remember, RNases ARE STILL EVERYWHERE!! Use the barrier pipet tips, wear gloves, and close the caps of all tubes.
Remember, RNases ARE STILL EVERYWHERE!! Use the barrier pipet tips, wear gloves, and close the caps of all tubes.
- Set up a RNA-RT primer mix for EACH RNA
sample (4 TOTAL = 2 control + 2 experimental) in 0.5 ml RNase-free
tubes:
1-5 µl TOTAL RNA (0.5-2.5 µg)
1 µl RT primer (use Cy3 for the control sample; use
Cy5 for the experimental sample)
nuclease-free water to a FINAL volume of 6 µl
- Mix by gently tapping tube; pulse spin.
- Heat to 80°C for 5 minutes (use the thermal cycler block); IMMEDIATELY transfer to ICE for 2-3 minutes; pulse spin and return to ice.
- Prepare a Reaction Master Mix:
- 10 µl 5X SuperScript II First Strand Buffer
- 5 µl 0.1M DTT
- 2.5 µl Superase-in RNase inhibitor
- 2.5 µl dNTP mix
- 2.5 µl SuperScript II reverse transcriptase (Invitrogen, 200U/µl)
Gently mix by tapping and pulse spin; keep on ice until ready to use.
- Add 4.5 µl of the Reaction Master Mix to the 6 µl RNA-RT primer mix (10.5 µl FINAL volume).
- Mix by gently tapping tube; pulse spin.
- Incubate at 42°C for 2 hours (use the thermal cycler block).
 During this incubation, practice adding liquid to the coverslip with lifters:
During this incubation, practice adding liquid to the coverslip with lifters:
Use a KimWipe to remove dust from a clear glass slide; center
a 25x60mm coverslip, LIFTERS-SIDE DOWN, on the
slide; very SLOWLY and CAREFULLY pipet 60 µl RO
water at the edge of the coverslip; make sure that
you use sterile WHITE tips, NOT the bulk YELLOW ones.
NOTE: capillary action will pull the liquid under the coverslip; you should see it evenly spread until it reaches the other end of the coverslip.
**The loading must be even and without any bubbles so that the entire array will be exposed to the labeled samples.
{Loading the microarray is very similar to casting the IEF gel in Protein Lab--just on a much smaller scale.}
 We have cDNA! Breathe...Relax a little...but still wear gloves and keep tubes closed. NOW we can use regular autoclaved pipet tips.
We have cDNA! Breathe...Relax a little...but still wear gloves and keep tubes closed. NOW we can use regular autoclaved pipet tips.
- STOP the RT reaction by adding 1 µl 1 M NaOH/100 mM EDTA.
- Incubate at 65°C for 10 minutes (use the thermal cycler block) to denature the DNA/RNA hybrids and degrade the RNA.
- Pulse spin the samples.
- NEUTRALIZE the reaction by adding 1.2 µl 2 M Tris-HCl, pH 7.5 (12.7 µl FINAL volume).
- Store cDNA synthesis reactions at -20°C until ready to proceed with cDNA Hybridization.
Copyright, Acknowledgements,
and Intended Use
Created by B. Beason (bbeason@rice.edu), Rice University, 5 June 2002
Updated 7 March 2007

 Home
Home