Data used in the Chemical Engineering modules has been stored in matrices. Since matrices must be rectangular and can hold either numeric or character data, this placed limits on the way the data was stored during use in the programs. It also made it rather difficult to extract data from the property data files and put the data in the arrays where they would be used. In Matlab versions through 4.2 this was the only choice for storage of data. Now Matlab 5.1 offers several choices that have definite advantages since they can store mixed (both numeric and character) data, can store data in higher dimensional arrays than matrices and can be used to avoid having to spend as much effort in storing and extracting the data from files. This chapter will show the basic ideas that were used in utilizing structures in the start301 program developed by Mike Cohen during the Summer of 1998.
First let's a brief review of the way chemical property data is stored in a chemical engineering session. The main types of arrays are:
|
|
|
|
|
|
|
|
|
4 |
|
|
|
|
ammonia nitrogen hydrogen argon |
|
|
|
|
17.0300
28.0130
2.0160
39.9480
|
|
|
|
|
15.494 13.45 12.78 13.915 2363.20 658.22 232.320 832.78 -22.62 -2.854 8.08 2.36 |
Cells could be used to store all four types of data in the same
array. We will give an example where data for the same four compounds
in Table 13.1 were put in a file with start301a and loaded with
start301b (these are the programs that start301
replaced. We see the familiar listing of compounds and
reactions at the end of this process:
Here are your compounds' formulae and names: No. Formula Name 1 NH3 ammonia 2 N2 nitrogen 3 H2 hydrogen 4 Ar argon Here are your reactions: 1 N2 + 3 H2 --> 2 NH3
We can consturct an empty cell array as indicated in the help notes
about the command by that name:
»help cell
CELL Create cell array.
CELL(N) is an N-by-N cell arrray of empty matrices.
CELL(M,N) or CELL([M,N]) is an M-by-N cell array of empty
matrices.
CELL(M,N,P,...) or CELL([M N P ...]) is an M-by-N-by-P-by-...
cell array of empty matrices.
CELL(SIZE(A)) is a cell array the same size as A containing
all empty matrices.
Thus we can create a cell to hold the data shown in Table 13.1 by:
»celldat=cell(4,1) <-- Making a empty, cell, column vector with place
celldat = for four "things".
[]
[]
[]
[]
»celldat{1,1}=nc <-- Putting nc in the first place.
Note use of "{" and "}"
celldat =
[4]
[]
[]
[]
»celldat{2,1}=cnms; <-- Adding cnms in the second place.
»celldat{3,1}=mw; <-- mw is put in the third place
»celldat{4,1}=Aabc; <-- The matrix Aabc is put in the fourth place.
»celldat <-- When we list the cell, we see the data we placed
celldat = only in the scalar: nc
[ 4] <-- nc is 4
[4x9 char ] <-- cnms is a 4 by 9 character matrix.
[4x1 double] <-- mw is a numeric column vector with 4 elements
[4x3 double] <-- Aabc is a 4 by 3 numeric matrix
The use of braces ( "{" and "}") to access the contents of a cell must be remembered. If you use parentheses instead you will find that it can lead to confusion:
»celldat(1,1) <-- This produces a cell with the number 4 in it
ans =
[4]
»celldat{1,1} <-- This gives us the number 4
ans =
4
»celldat(4,1) <-- A cell having Aabc in it
ans =
[4x3 double]
»celldat{4,1} <-- The numeric values of the coefficients
ans =
1.0e+03 *
0.0155 2.3632 -0.0226
0.0134 0.6582 -0.0029
0.0128 0.2323 0.0081
0.0139 0.8328 0.0024
The same data may also be placed in a structure. Here is information to get started:
»help struct
STRUCT Create or convert to structure array.
S = STRUCT('field1',VALUES1,'field2',VALUES2,...)
creates a structure array with the specified fields and values. The value arrays VALUES1, VALUES2, etc. must be cell arrays of the same size, scalar cells or single values. Corresponding elements of the value arrays are placed into corresponding structure array elements.
The size of the resulting structure is the same size as the value cell arrays or 1-by-1 if none of the values is a cell.
STRUCT(OBJ)
converts the object OBJ into its equivalent structure. The class information is lost.
Example:
s = struct('type',{'big','little'},'color','red','x',{3 4})
See also CLASS, CELL, GETFIELD, SETFIELD, RMFIELD, FIELDNAMES.
From this help, we can make a structure with the same data we put in celldat:
»strucdat=struct('nc',nc,'cnms',cnms,'mw',mw,'Aabc',Aabc)
strucdat =
nc: 4
cnms: [4x9 char ]
mw: [4x1 double]
Aabc: [4x3 double]
Now if we want to access this data that we stored, we can use:
»strucdat.nc
ans =
4
»strucdat.mw
ans =
17.0300
28.0130
2.0160
39.9480
Matlab structures seem to offer the most efficient way to store and then retreive data. The main advantages of structures over cells are:
Two functions and a starting data base with part of the data in our data files were developed to explore the possibility of replacing our starting programs start301a and start301b with a single Matlab program. Help applied to the commands: fieldnames, getfield and setfield was used to understand ways that this might be done. The example programs: mkstruct and mkvars illustrate some of the advantages of using structures.
FIELDNAMES Get structure field names.
NAMES = FIELDNAMES(S)
returns the structure field names associated with the structure S as a cell array of strings.
See also GETFIELD, SETFIELD.
You must be careful in using fieldnames since it returns a
cell array.
>> fnames=fieldnames(strucdat)
fnames =
'nc'
'cnms'
'mw'
'Aabc'
The class command tells us that:
>> class(fnames) ans = cell
One element of fnames may look like a string, but it is not.
>> class(fnames)
ans =
cell
>> fn1=fnames(1)
fn1 =
'nc'
>> 'nc'==fn1
??? Function '==' not defined for variables of class 'cell'.
>> class(fn1)
ans =
cell
If we want the contents of a cell, we need to use braces:
>> fn1=fnames{1}
fn1 =
nc
>> 'nc'==fn1
ans =
1 1
>> class(fn1)
ans =
char
GETFIELD Get structure field contents.
F = GETFIELD(S,'field')
returns the contents of the specified field. This is equivalent to the syntax F = S.field.
S must be a 1-by-1 structure.
F = GETFIELD(S,{i,j},'field',{k})
is equivalent to the syntax
F = S(i,j).field(k).
In other words,
F = GETFIELD(S,sub1,sub2,...)
returns the contents of the structure S using the subscripts or field references specified in sub1,sub2,etc. Each set of subscripts in parentheses must be enclosed in a cell array and passed to GETFIELD as a separate input. Field references are passed as strings.
See also SETFIELD, FIELDNAMES.
SETFIELD Set structure field contents.
S = SETFIELD(S,'field',V)
sets the contents of the specified field to the value V. This is equivalent to the syntax S.field = V. S must be a 1-by-1 structure. The changed structure is returned.
S = SETFIELD(S,{i,j},'field',{k},V)
is equivalent to the syntax
S(i,j).field(k) = V;
In other words,
S = SETFIELD(S,sub1,sub2,...,V)
sets the contents of the structure S to V using the subscripts or field references specified in sub1,sub2,etc. Each set of subscripts in parentheses must be enclosed in a cell array and passed to SETFIELD as a separate input. Field references are passed as strings.
See also GETFIELD, FIELDNAMES.
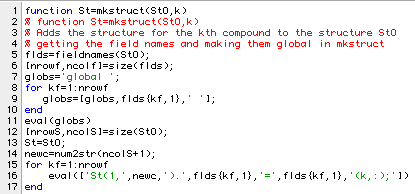
The function mkstruct allows us to add data for one compound to an existing structure. The fields in the existing structure are identical to the names of data variables set by start301a and start301b. Here is a listing of the program.

The function mkvars extracts all the data in a structure like the one created with mkstruct and puts the data in variables of the same names as the fields in the structure. All the compounds are added to these variables. Here is a listing of mkvars.
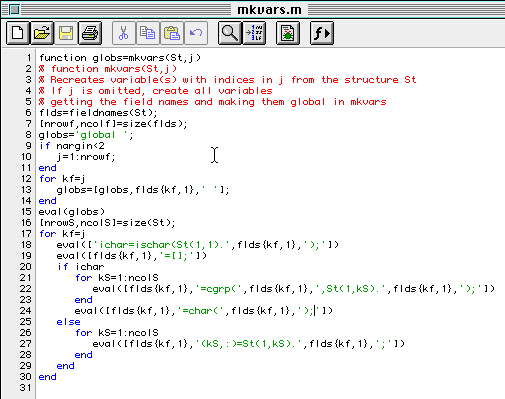
Note that the mkvars function returns a string with the command that makes all the variables global. Thus if this returned string is evaled, we will have a session ready to be used in the 301 modules.
The function mkstruct was used to produce a structure holding twelve types of data for twenty compounds. This procedure produced a structure called stdat. It was saved as the file: /home/ceng301/datbas/struct1.mat. Here is an example of using the mkstruct function to add the compound ethanol to the structure. First we use start301a and start301b:
»start301b Copyright 1996 Rice University All rights reserved If you have not run the FORTRAN program start301a to produce a data file, do so now or you can just set the names without any data. A blank reply for the file name will let you just set the names. Give the name of your data file:waterethanol How many streams will there be?1 Here are your compounds' formulae and names: No. Formula Name 1 H2O water 2 C2H5OH ethanol
Next we load the file that had data for our twenty compounds:
»load struct1 »who Your variables are: Aabc StStdh cpl form icpv stdat AcF StdhkJ cps hcpl mw Gibb TbpK cpv hcps nc LJones Tdeg critP hcpv ne LhlvkJ TmpK critT icpl ns LhslkJ cnms critZ icps nst
The structure stdat has the form shown by:
»stdat
stdat =
20x1 struct array with fields:
cnms
form
mw
Aabc
LhlvkJ
StStdh
StdhkJ
TbpK
cpl
cpv
hcpl
hcpv
Here is the data for the first compound in the structure:
»stdat(1)
ans =
cnms: ' benzene '
form: ' C6H6 '
mw: 78.1130
Aabc: [14.1600 2.9488e+03 -44.5630]
LhlvkJ: 30.7600
StStdh: 2
StdhkJ: 82.9300
TbpK: 353.2600
cpl: [-7.2732 0.7705 -0.0016 1.8979e-06]
cpv: [18.5800 -0.0117 0.0013 -2.0790e-06 1.0500e-09]
hcpl: [4.7448e-10 -5.4937e-07 3.8527e-04 -0.0073 28.4482]
hcpv: [2.1000e-13 -5.1975e-10 4.2500e-07 -5.8700e-06 0.0186 70.2605]
Let's add the ethanol data to this data structure:
»stdat2=mkstruct(stdat',2) <-- Note transpose of stdat
stdat2 =
1x21 struct array with fields:
cnms
form
mw
Aabc
LhlvkJ
StStdh
StdhkJ
TbpK
cpl
cpv
hcpl
hcpv
»stdat2(21)<-- Seeing what was added for ethanol
ans =
cnms: ' ethanol'
form: ' C2H5OH'
mw: 46.0690
Aabc: [16.1950 3.4235e+03 -55.7150]
LhlvkJ: 38.5800
StStdh: 2
StdhkJ: -234.8000
TbpK: 351.4800
cpl: [-325.1300 4.1379 -0.0140 1.7035e-05]
cpv: [17.6900 0.1495 8.9480e-05 -1.9730e-07 8.3170e-11]
hcpl: [4.2588e-09 -4.6767e-06 0.0021 -0.3251 -272.9021]
hcpv: [1.6634e-14 -4.9325e-11 2.9827e-08 7.4750e-05 0.0177 -247.1590]
Let's save this new structure and reload it into a clear workspace, then use mkvars create a new set of variables for use in the 301 modules.
»save structnew stdat2 »clear »load structnew »who Your variables are: stdat2
Now use mkvars:
»mkvars(stdat2)
ans =
global cnms form mw Aabc LhlvkJ StStdh StdhkJ TbpK cpl cpv hcpl hcpv
»eval(ans) <-- This makes the variables global (you could also do
»who do it by: eval(mkvars(stdat2)).
Your variables are:
Aabc StdhkJ cnms form mw
LhlvkJ TbpK cpl hcpl stdat2
StStdh ans cpv hcpv
»cnms <-- We can now look at all the compounds in struct1 plus
ethanol that we added in the session.
cnms =
benzene
n-butane
n-butanol
n-butyric acid
n-heptane
n-hexadecane
n-hexane
n-octane
n-pentane
n-propanol
neon
nicotinic acid
nitric acid
nitric oxide
nitrogen
nitrogen dioxide
nitromethane
nitrous oxide
toluene
water
ethanol
»Aabc <-- checking to see that one of the numeric arrays is also
set correctly.
Aabc =
1.0e+03 *
0.0142 2.9488 -0.0446
0.0140 2.2924 -0.0279
0.0147 2.9030 -0.1029
0.0158 4.0961 -0.0718
0.0139 2.9327 -0.0556
0.0142 4.2053 -0.1192
0.0141 2.8254 -0.0427
0.0142 3.3042 -0.0552
0.0140 2.5546 -0.0363
0.0152 3.0083 -0.0865
NaN NaN NaN
NaN NaN NaN
NaN NaN NaN
0.0169 1.3191 -0.0141
0.0134 0.6582 -0.0029
NaN NaN NaN
NaN NaN NaN
NaN NaN NaN
0.0143 3.2424 -0.0472
0.0165 3.9854 -0.0390
0.0162 3.4235 -0.0557
»